6. Tidy Data#
6.1. Introduction#
In this chapter, you will learn a consistent way to organise your data in Python using the principle known as tidy data. Tidy data is not appropriate for everything, but for a lot of analysis and a lot of tabular data it will be what you need. Getting your data into this format requires some work up front, but that work pays off in the long term. Once you have tidy data, you will spend much less time munging data from one representation to another, allowing you to spend more time on the data questions you care about.
In this chapter, you’ll first learn the definition of tidy data and see it applied to simple toy dataset. Then we’ll dive into the main tool you’ll use for tidying data: melting. Melting allows you to change the form of your data, without changing any of the values. We’ll finish up with a discussion of usefully untidy data, and how you can create it if needed.
If you particularly enjoy this chapter and want to learn more about the underlying theory, you can learn more in the Tidy Data paper published in the Journal of Statistical Software.
6.1.1. Prerequisites#
This chapter will use the pandas data analysis package.
6.2. Tidy Data#
There are three interrelated features that make a dataset tidy:
Each variable is a column; each column is a variable.
Each observation is row; each row is an observation.
Each value is a cell; each cell is a single value.
The figure below shows this:

Why ensure that your data is tidy? There are two main advantages:
There’s a general advantage to picking one consistent way of storing data. If you have a consistent data structure, it’s easier to learn the tools that work with it because they have an underlying uniformity. Some tools, for example data visualisation package seaborn, are designed with tidy data in mind.
There’s a specific advantage to placing variables in columns because it allows you to take advantage of pandas’ vectorised operations (operations that are more efficient).
Tidy data aren’t going to be appropriate every time and in every case, but they’re a really, really good default for tabular data. Once you use it as your default, it’s easier to think about how to perform subsequent operations.
Having said that tidy data are great, they are, but one of pandas’ advantages relative to other data analysis libraries is that it isn’t too tied to tidy data and can navigate awkward non-tidy data manipulation tasks happily too.
There are two common problems you find in data that are ingested that make them not tidy:
A variable might be spread across multiple columns.
An observation might be scattered across multiple rows.
For the former, we need to “melt” the wide data, with multiple columns, into long data.
For the latter, we need to unstack or pivot the multiple rows into columns (ie go from long to wide.)
We’ll see both below.
6.3. Tools to Make Data Tidy with pandas#
6.3.1. Melt#
melt() can help you go from “wider” data to “longer” data, and is a really good one to remember.
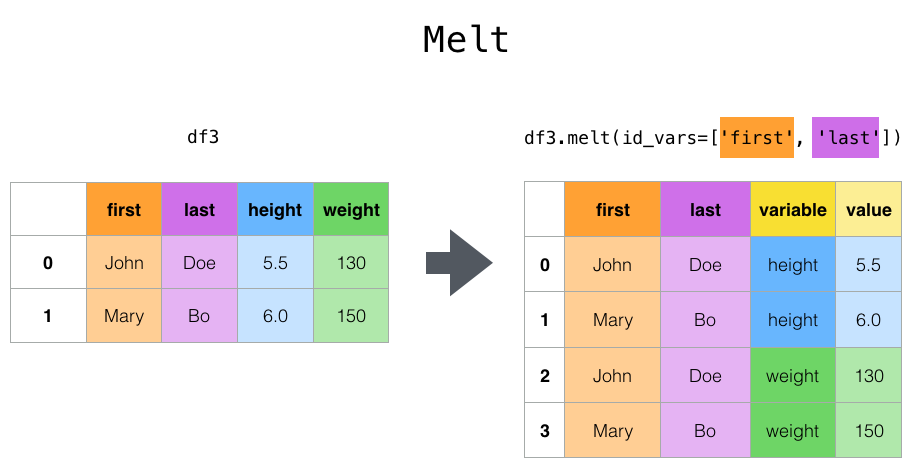
Here’s an example of it in action:
import pandas as pd
df = pd.DataFrame(
{
"first": ["John", "Mary"],
"last": ["Doe", "Bo"],
"job": ["Nurse", "Economist"],
"height": [5.5, 6.0],
"weight": [130, 150],
}
)
print("\n Unmelted: ")
print(df)
print("\n Melted: ")
df.melt(id_vars=["first", "last"], var_name="quantity", value_vars=["height", "weight"])
Unmelted:
first last job height weight
0 John Doe Nurse 5.5 130
1 Mary Bo Economist 6.0 150
Melted:
| first | last | quantity | value | |
|---|---|---|---|---|
| 0 | John | Doe | height | 5.5 |
| 1 | Mary | Bo | height | 6.0 |
| 2 | John | Doe | weight | 130.0 |
| 3 | Mary | Bo | weight | 150.0 |
Exercise
Perform a melt() that uses job as the id instead of first and last.
How does this relate to tidy data? Sometimes you’ll have a variable spread over multiple columns that you want to turn tidy. Let’s look at this example that uses cases of tuburculosis from the World Health Organisation.
First let’s open the data and look at the top of the file.
df_tb = pd.read_parquet(
"https://github.com/aeturrell/python4DS/raw/refs/heads/main/data/who_tb_cases.parquet"
)
df_tb.head()
| country | 1999 | 2000 | |
|---|---|---|---|
| 0 | Afghanistan | 745.0 | 2666.0 |
| 1 | Brazil | 37737.0 | 80488.0 |
| 2 | China | 212258.0 | 213766.0 |
You can see that we have two columns for a single variable, year. Let’s now melt this.
df_tb.melt(
id_vars=["country"],
var_name="year",
value_vars=["1999", "2000"],
value_name="cases",
)
| country | year | cases | |
|---|---|---|---|
| 0 | Afghanistan | 1999 | 745.0 |
| 1 | Brazil | 1999 | 37737.0 |
| 2 | China | 1999 | 212258.0 |
| 3 | Afghanistan | 2000 | 2666.0 |
| 4 | Brazil | 2000 | 80488.0 |
| 5 | China | 2000 | 213766.0 |
We now have one observation per row, and one variable per column: tidy!
6.3.2. A simpler wide to long#
If you don’t want the headscratching of melt(), there’s also wide_to_long(), which is really useful for typical data cleaning cases where you have data like this:
import numpy as np
df = pd.DataFrame(
{
"A1970": {0: "a", 1: "b", 2: "c"},
"A1980": {0: "d", 1: "e", 2: "f"},
"B1970": {0: 2.5, 1: 1.2, 2: 0.7},
"B1980": {0: 3.2, 1: 1.3, 2: 0.1},
"X": dict(zip(range(3), np.random.randn(3))),
"id": dict(zip(range(3), range(3))),
}
)
df
| A1970 | A1980 | B1970 | B1980 | X | id | |
|---|---|---|---|---|---|---|
| 0 | a | d | 2.5 | 3.2 | -0.617958 | 0 |
| 1 | b | e | 1.2 | 1.3 | -0.286581 | 1 |
| 2 | c | f | 0.7 | 0.1 | -1.478036 | 2 |
i.e. data where there are different variables and time periods across the columns. Wide to long is going to let us give info on what the stubnames are (‘A’, ‘B’), the name of the variable that’s always across columns (here, a year), any values (X here), and an id column.
pd.wide_to_long(df, stubnames=["A", "B"], i="id", j="year")
| X | A | B | ||
|---|---|---|---|---|
| id | year | |||
| 0 | 1970 | -0.617958 | a | 2.5 |
| 1 | 1970 | -0.286581 | b | 1.2 |
| 2 | 1970 | -1.478036 | c | 0.7 |
| 0 | 1980 | -0.617958 | d | 3.2 |
| 1 | 1980 | -0.286581 | e | 1.3 |
| 2 | 1980 | -1.478036 | f | 0.1 |
6.3.3. Stack and Unstack#
Stack, stack() is a shortcut for taking a single type of wide data variable from columns and turning it into a long form dataset, but with an extra index.
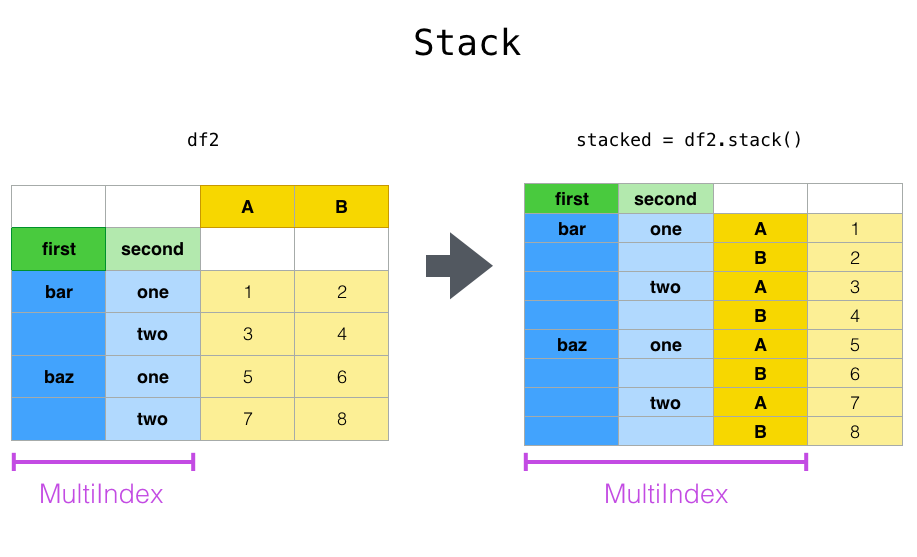
Unstack, unstack() unsurprisingly does the same operation, but in reverse.
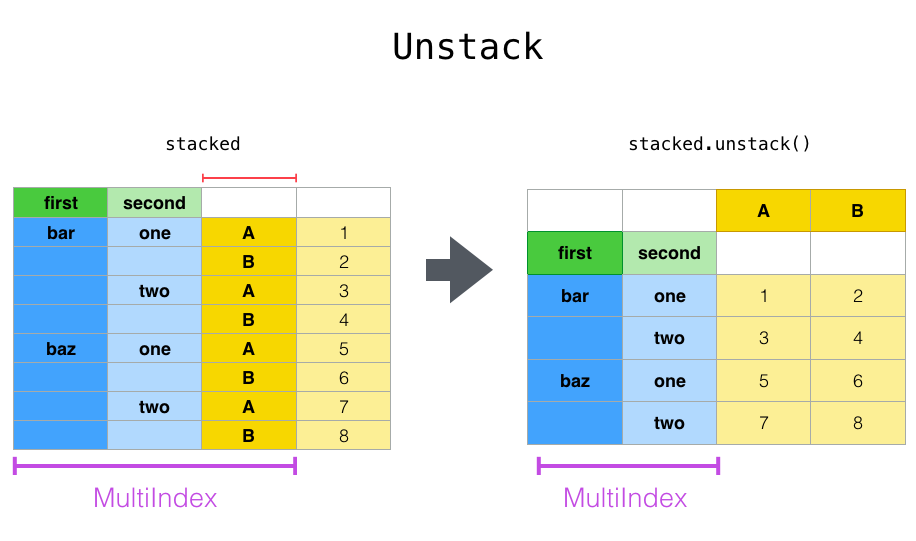
Let’s define a multi-index data frame to demonstrate this:
tuples = list(
zip(
*[
["bar", "bar", "baz", "baz", "foo", "foo", "qux", "qux"],
["one", "two", "one", "two", "one", "two", "one", "two"],
]
)
)
index = pd.MultiIndex.from_tuples(tuples, names=["first", "second"])
df = pd.DataFrame(np.random.randn(8, 2), index=index, columns=["A", "B"])
df
| A | B | ||
|---|---|---|---|
| first | second | ||
| bar | one | -0.730825 | 1.282711 |
| two | 0.724691 | 0.953490 | |
| baz | one | -0.559218 | -2.131779 |
| two | 1.005972 | 0.231563 | |
| foo | one | -1.479746 | 0.311247 |
| two | -0.423248 | 1.883557 | |
| qux | one | -2.255293 | 1.028665 |
| two | -0.021046 | -0.140835 |
Let’s stack this to create a tidy dataset:
df = df.stack()
df
first second
bar one A -0.730825
B 1.282711
two A 0.724691
B 0.953490
baz one A -0.559218
B -2.131779
two A 1.005972
B 0.231563
foo one A -1.479746
B 0.311247
two A -0.423248
B 1.883557
qux one A -2.255293
B 1.028665
two A -0.021046
B -0.140835
dtype: float64
This has automatically created a multi-layered index but that can be reverted to a numbered index using df.reset_index()
Now let’s see unstack but, instead of unstacking the ‘A’, ‘B’ variables we began with, let’s unstack the ‘first’ column by passing level=0 (the default is to unstack the innermost index). This diagram shows what’s going on:
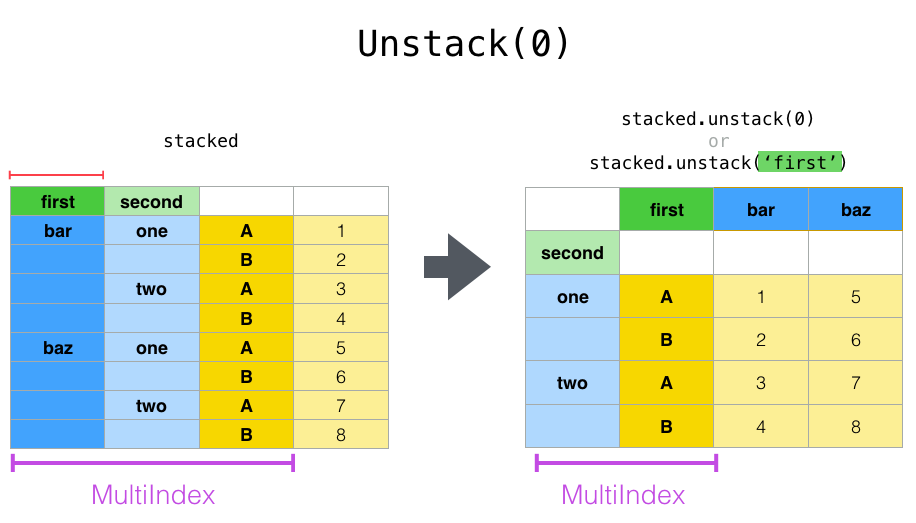
And here’s the code:
df.unstack(level=0)
| first | bar | baz | foo | qux | |
|---|---|---|---|---|---|
| second | |||||
| one | A | -0.730825 | -0.559218 | -1.479746 | -2.255293 |
| B | 1.282711 | -2.131779 | 0.311247 | 1.028665 | |
| two | A | 0.724691 | 1.005972 | -0.423248 | -0.021046 |
| B | 0.953490 | 0.231563 | 1.883557 | -0.140835 |
Exercise
What happens if you unstack to level=1 instead? What about applying unstack() twice?
6.3.4. Pivoting data from long to wide#
pivot() and pivot_table() help you to sort out data in which a single observation is scattered over multiple rows.
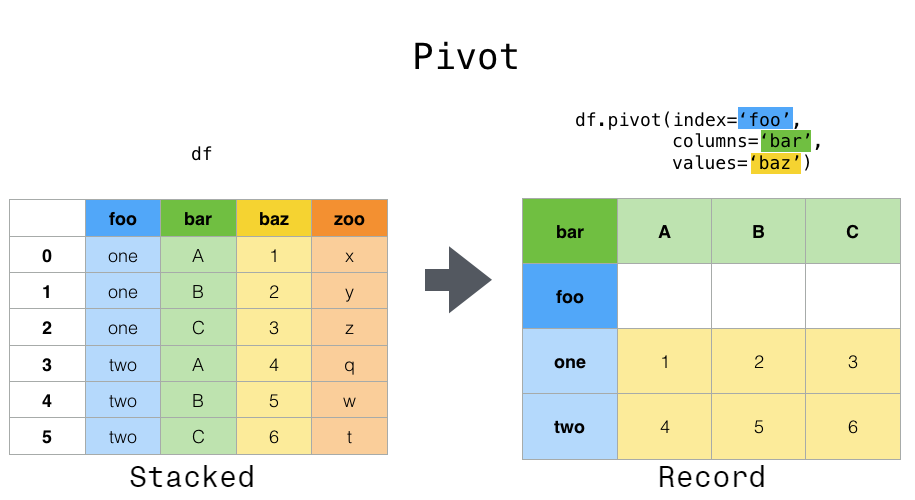
Here’s an example dataframe where observations are spread over multiple rows:
df_tb_cp = pd.read_parquet(
"https://github.com/aeturrell/python4DS/raw/refs/heads/main/data/who_tb_case_and_pop.parquet"
)
df_tb_cp.head()
| country | year | type | count | |
|---|---|---|---|---|
| 0 | Afghanistan | 1999-01-01 | cases | 745 |
| 1 | Afghanistan | 1999-01-01 | population | 19987071 |
| 2 | Afghanistan | 2000-01-01 | cases | 2666 |
| 3 | Afghanistan | 2000-01-01 | population | 20595360 |
| 4 | Brazil | 1999-01-01 | cases | 37737 |
You see that we have, for each year-country, “case” and “population” in different rows.
Now let’s pivot this to see the difference:
pivoted = df_tb_cp.pivot(
index=["country", "year"], columns=["type"], values="count"
).reset_index()
pivoted
| type | country | year | cases | population |
|---|---|---|---|---|
| 0 | Afghanistan | 1999-01-01 | 745 | 19987071 |
| 1 | Afghanistan | 2000-01-01 | 2666 | 20595360 |
| 2 | Brazil | 1999-01-01 | 37737 | 172006362 |
| 3 | Brazil | 2000-01-01 | 80488 | 174504898 |
| 4 | China | 1999-01-01 | 212258 | 1272915272 |
| 5 | China | 2000-01-01 | 213766 | 1280428583 |
Pivots are especially useful for time series data, where operations like shift() or diff() are typically applied assuming that an entry in one row follows (in time) from the one above. When we do shift() we often want to shift a single variable in time, but if a single observation (in this case a date) is over multiple rows, the timing is going go awry. Let’s see an example.
import numpy as np
data = {
"value": np.random.randn(20),
"variable": ["A"] * 10 + ["B"] * 10,
"date": (
list(pd.date_range("1/1/2000", periods=10, freq="ME"))
+ list(pd.date_range("1/1/2000", periods=10, freq="ME"))
),
}
df = pd.DataFrame(data, columns=["date", "variable", "value"])
df.sample(5)
| date | variable | value | |
|---|---|---|---|
| 12 | 2000-03-31 | B | -0.055590 |
| 1 | 2000-02-29 | A | 1.272996 |
| 10 | 2000-01-31 | B | -0.704969 |
| 17 | 2000-08-31 | B | -0.991378 |
| 5 | 2000-06-30 | A | 0.451136 |
If we just run shift() on the above, it’s going to shift variable B’s and A’s together even though these overlap in time and are different variables. So we pivot to a wider format (and then we can shift in time safely).
df.pivot(index="date", columns="variable", values="value").shift(1)
| variable | A | B |
|---|---|---|
| date | ||
| 2000-01-31 | NaN | NaN |
| 2000-02-29 | 0.125933 | -0.704969 |
| 2000-03-31 | 1.272996 | 0.228826 |
| 2000-04-30 | -0.040630 | -0.055590 |
| 2000-05-31 | 0.135487 | 0.965950 |
| 2000-06-30 | 0.507748 | 0.537605 |
| 2000-07-31 | 0.451136 | -0.113129 |
| 2000-08-31 | 1.012889 | -1.198560 |
| 2000-09-30 | -0.234788 | -0.991378 |
| 2000-10-31 | 0.441697 | -0.918995 |
Exercise
Why is the first entry NaN?
Exercise
Perform a pivot() that applies to both the variable and category columns in the example from above where category is defined such that `df[“category”] = np.random.choice([“type1”, “type2”, “type3”, “type4”], 20). (Hint: remember that you will need to pass multiple objects via a list.)


